How is population decline influenced by evolutionary mechanisms?
We are interested in studying what causes populations to decline. Variation between populations is driven by a number of evolutionary mechanisms, for example, the availability of prey species, introduction of disease, inbreeding depression and intraspecific phenotypic variation. Evaluating the ecological and evolutionary interactions within populations is important for informing effective conservation management of endemic species.
To conserve endemic species in Aotearoa, we can use population genetics tools to characterise population structure, estimate population sizes, and identify local adaptation. When informing conservation management efforts, understanding evolutionary mechanisms is important to get the full picture of factors impacting population persistence. The Flanagan lab is exploring non-invasive molecular methods for assessing Pōhatu’s kororā population’s genetic variation. We are particularly interested in breeding pairs where parents are a mix of the white-flippered and little blue morphotypes. We hope to use molecular methods to better understand the persistence of the white-flippered morphotype, which is unique to the Canterbury region, and explore the impacts phylogenetic constraints may be having on the colony. Additionally, we are working on modelling the distribution of the wide-bodied pipefish, Stigmatopora nigra, and using genetic analysis to compare population structure across its range. By comparing phenotypic and genotypic data of S. nigra across Aotearoa we will evaluate different selection pressures, assess the evolutionary mechanisms that drive local populations, and expand on the currently limited understanding of S. nigra.
Understanding factors of population decline is critical for conserving the species they affect. The Flanagan lab is currently focused on diet and disease as agents of decline. In collaboration with Mana Whenua, Pohatu Penguins and the LaRue and ConSERT labs we are utilizing long-read nanopore sequencing to assess the diet of the Pohatu population of kororā as a potential driver of local population decline. Using non-invasive fecal and molt feather samples to develop a baseline understanding of local diet, this project aims to facilitate community engagement in genetic sampling of local populations. Collaboration with the Banks Peninsula community will be further supported through the development of remote sequencing using the portable MinION device (Oxford nanopore technologies). In addition to diet in kororā populations, the Flanagan lab is interested in the coevolution of invasive diseases, specifically Phytophthora, and the plants they impact. In collaboration with Claudia Meisrimler, we are looking at Phytophthora cinnamomi, which causes dieback non-discriminantly across endemic and economically important flora and is a close relative to P. agathidicida, the source of Kauri dieback. Improving our understanding of the P. cinnamoni genome and how it co-evolves with crop species will allow for better informed management and conservation programmes aimed at tackling dieback diseases.

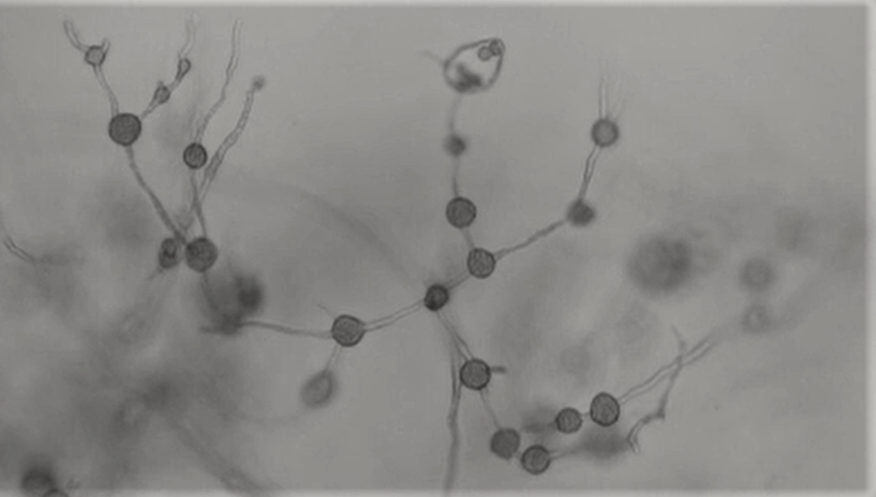 Phytophthora Pluvialis. Credit Sophie Eccersall
Phytophthora Pluvialis. Credit Sophie Eccersall
Some relevant papers include:
Howell LA, LaRue M, Flanagan SP (2021) Environmental DNA as a tool for monitoring Antarctic vertebrates. New Zealand Journal of Zoology. DOI: 10.1080/03014223.2021.1900299. link to paper, PDF, my summary
Flanagan SP, and Jones AG. 2019. The future of parentage analysis: Moving from microsatellites to SNPs and beyond. Molecular Ecology. 28(3): 544-567 doi: 10.1111/mec.14988 link to paper, my summary
Jost L, Archer F, Flanagan SP, Gaggiotti O, Hoban S, Latch E. 2018. Differentiation measures for conservation genetics. Evolutionary Applications. 11: 1139-1148. doi: 10.1111/eva.12590. pdf, link to paper, my summary
, , , , . 2017. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evolutionary Applications. 11: 1035–1052. doi: 10.1111/eva.12569. pdf, link to paper, my summary